Annex II – Examples of European patent applications
|
A revised version of this publication entered into force. |
METHOD FOR DETERMINING TOLERANCE OF CANCER CELL TO EPIDERMAL GROWTH FACTOR RECEPTOR INHIBITOR
Title of invention (designation in request for grant suffices)
Technical Field
The present invention relates to a method for determining tolerance of a cancer cell to an epidermal growth factor receptor inhibitor (hereinafter will be referred to as "EGFR inhibitor") in a human patient suffering from a cancer.
R. 42(1)(a)
Technical field to which invention relates
Background Art
EGFR inhibitors, for example, anti-epidermal growth factor receptor antibody drugs (anti-EGFR antibody drug) such as cetuximab and panitumumab have been known as a therapeutic agent for cancer. These EGFR inhibitors act in cancer cells to inhibit the function of an epidermal growth factor receptor (EGFR) which is involved in cancer cell growth.
R. 42(1)(b)
Relevant prior art
Meanwhile, it has been known that, in a case where cancer cells of a cancer patient have a mutation in the KRAS gene, the cancer cells are resistant to the EGFR inhibitor, and therefore effects of the EGFR inhibitor are reduced. The EGFR inhibitor has side effects such as skin disorder, and therefore it is preferable to administer the EGFR inhibitor only to cancer patients for whom high therapeutic efficacy may be expected by administration of the EGFR inhibitor. As a method for determining whether a treatment with the EGFR inhibitor is effective or not to the patient, a method for checking the presence or absence of a mutation in the KRAS gene of the cancer cells of the patient has been known.
For example, WO2014/148557 discloses a method for predicting sensitivity to EGFR inhibitors, in which, in a case where a nucleic acid derived from a mutant-type KRAS gene or a protein thereof is detected in a blood sample, it is determined that a possibility that a tumour is not sensitive to the EGFR inhibitor is high.
Assessment of prior art
According to de Roock et al, Lancet Oncology, vol. 12, 2011, p. 594 – 603, BRAF mutation V600E confers resistance of cancer patients to EGFR antibodies.
Technical Problem
The inventors of the present invention have found that the treatment with the EGFR inhibitor is not effective in some cases even to the cancer patient whose cancer cells have the wild-type KRAS gene.
R. 42
Technical problem
The present invention has been made in view of the above problems, and an object of the present invention is to select in advance a cancer patient for whom a treatment with the EGFR. inhibitor is ineffective.
Solution to Problem
The present invention is a method for determining tolerance of a cancer cell to an EGFR inhibitor in a human patient suffering from a cancer, the method comprising a step of determining the presence or absence of a mutation of a 326th amino acid residue of an amino acid sequence of a B-Raf protein (hereinafter referred to as "B-Raf") of the cancer cell by using a sample containing the cancer cell collected from the human patient, wherein the cancer cell is determined as tolerant to the epidermal growth factor receptor inhibitor, when the mutation of the amino acid residue is present.
R. 42(1)(c)
Disclosure of invention
R. 42(1)(c)
Advantageous effects of invention
According to this method, it is possible to determine the tolerance of the cancer cell to the EGFR inhibitor by determining the presence or absence of the mutation of a specific amino acid residue, which is the 326th amino acid residue, in the amino acid sequence of the B-Raf of the cancer cell, and therefore it is possible to select in advance a cancer patient for whom a treatment with the EGFR inhibitor is ineffective.
The above-described EGFR inhibitor may be an anti-epidermal growth factor receptor antibody drug.
The determination of the presence or absence of the mutation of the amino acid residue described above may include detection of a mutation of a base sequence encoding the 326th amino acid residue of the amino acid sequence of the B-Raf.
The detection of the mutation of the base sequence described above may be performed by DNA sequencing, polymerase chain reaction, allele-specific amplification, hybridization using allele-specific probes, mismatch cleavage analysis, single-strand conformation polymorphism, denaturing gradient gel electrophoresis, or temperature gradient gel electrophoresis.
The above-described sample may be a cancer resection tissue specimen, a biopsy specimen, an ascites-infiltrating cancer cell, a circulating cancer cell, serum, or plasma. If these samples are used as a sample, it is possible to select with higher certainty a cancer patient for whom the treatment with the EGFR inhibitor is ineffective.
The above-described cancer may be a colorectal cancer or a rectal cancer. If the cancer is a colorectal cancer or a rectal cancer, it is possible to select with higher certainty a cancer patient for whom the treatment with the EGFR inhibitor is ineffective.
The mutation of the amino acid residue described above may be I326V. If the mutation is I326V, it is possible to select with higher certainty a cancer patient for whom the treatment with the EGFR inhibitor is ineffective.
The mutation of the base sequence described above may be c.976A>G. If the mutation is c.976A>G it is possible to select with higher certainty a cancer patient for whom the treatment with the EGFR inhibitor is ineffective.
The above-described cancer cell preferably has a wild-type KRAS gene. With the cancer cell having the wild-type KRAS gene, it is possible to select with higher certainty a cancer patient for whom the treatment with the EGFR inhibitor is ineffective.
Advantageous Effects of Invention
According to the present invention, it is possible to select in advance a cancer patient for whom the treatment with the EGFR inhibitor is ineffective.
Advantageous effects of invention
Brief Description of Drawings
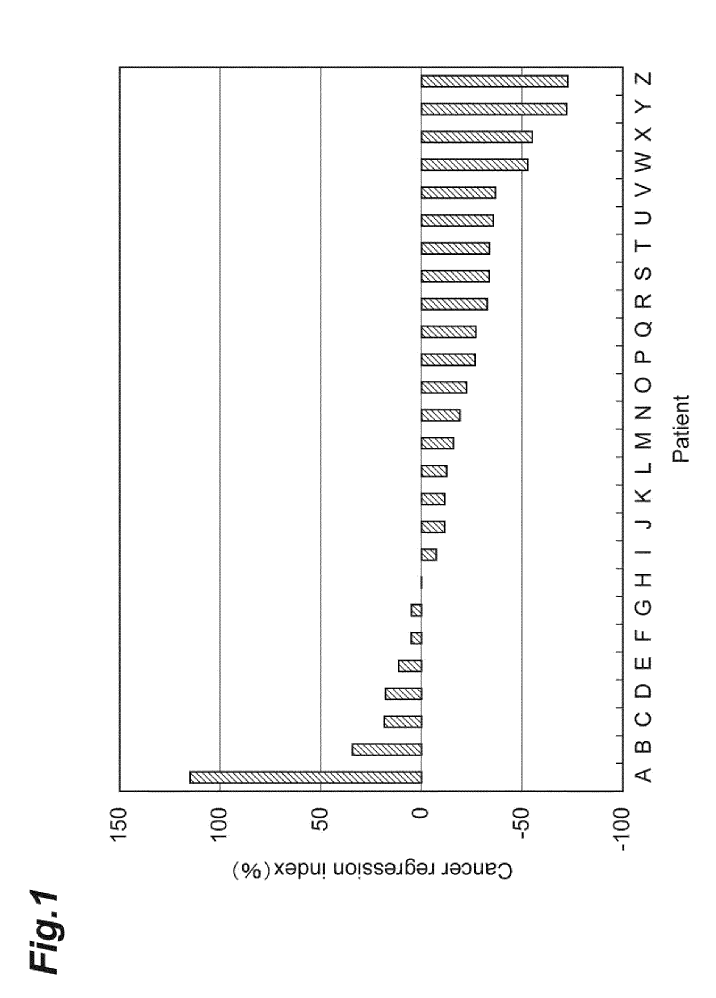
Fig. 1 is a graph showing results of an example.
R. 42(1)(d)
Brief description of drawings
Description of Embodiments
Hereinafter, embodiments for carrying out the present invention will be described in detail. However, the present invention is not limited to the following embodiments.
R. 42(1)(e)
Description of at least one way of carrying out the invention with reference to drawings
In one aspect, the present invention is a method for determining tolerance of a cancer cell to an EGFR inhibitor in a human patient suffering from a cancer (hereinafter referred to as "cancer patient"). The method according to this aspect comprises a step of determining the presence or absence of a mutation of a 326th amino acid residue of an amino acid sequence of a B-Raf of the cancer cell by using a sample containing the cancer cell collected from the human patient.
Examples of cancers for which the method according to the present invention may be used include colorectal cancer, rectal cancer, colon cancer, stomach cancer, liver cancer, thyroid cancer, uterine cancer, kidney cancer, pancreatic cancer, tongue cancer, prostate cancer, lung cancer, skin cancer, ovarian cancer, gallbladder cancer, head and neck cancer, testicular cancer, adrenal cancer, oral cancer, bone and soft tissue tumour, brain tumour, malignant melanoma, osteosarcoma, chondrosarcoma, rhabdomyosarcoma, leiomyosarcoma, leukaemia, malignant lymphoma, and multiple myeloma. Among these, in a case where the cancer is a colorectal cancer or a rectal cancer, it is possible to select with higher certainty a cancer patient for whom a treatment with the EGFR inhibitor is ineffective.
The "EGFR inhibitor" in the present description is not particularly limited as long as the EGFR inhibitor is a drug that inhibits expression or activity of the EGFR, and may be any of a low molecular compound targeting the EGFR, such as gefitinib and erlotinib, an anti-EGFR antibody drug, an EGFR antisense oligonucleotide, an aptamer, and the like. The anti-EGFR antibody drug is, for example, an antibody that inhibits binding of epidermal growth factor (EGF) to the EGFR. Examples of the anti-EGFR antibody drug include a monoclonal antibody that recognizes an extracellular domain of the EGFR as an epitope. Specific examples of the anti-EGFR antibody drug include cetuximab and panitumumab. The EGFR inhibitor may be used alone, or a plurality of EGFR inhibitors may be used in combination.
The "antibody" in the present description encompasses not only an antibody molecule having two complete light chains and two complete heavy chains, but also an antibody fragment capable of binding to an antigen. Examples of the antibody fragment include F(ab’)2, Fab’, Fab, and Fv. The antibody is preferably either a chimeric antibody, a humanized antibody, or a fully human antibody.
In addition, the method according to the present embodiment is effective also in a case of determining the tolerance of the cancer cell to a treatment in which the EGFR inhibitor and other anticancer agent are used in combination. Examples of the treatment in which the EGFR inhibitor and other anticancer agent are used in combination include CPT-11 + Panitumumab therapy, IRIS + Panitumumab therapy, FOLFOX (for example, mFOLFOX6) + Panitumumab therapy, FOLFIRI + Panitumumab therapy, CPT-11 + Cetuximab therapy, IRIS + Cetuximab therapy, FOLFOX (for example, mFOLFOX6) + Cetuximab therapy, FOLFIRI + Cetuximab therapy, and sLV5FU2 + Cetuximab therapy.
Examples of the sample containing the cancer cell include a cancer resection tissue specimen, a biopsy specimen, an ascites-infiltrating cancer cell, a circulating cancer cell, serum, plasma, blood, faeces, urine, sputum, cerebrospinal fluid, pleural fluid, nipple aspirate fluid, lymph fluid, cell culture liquid, and other tissues and fluids collected from the patient. From the viewpoint of selecting with higher certainty a cancer patient for whom a treatment with the EGFR inhibitor is ineffective, the sample containing the cancer cell is preferably a cancer resection tissue specimen, a biopsy specimen, an ascites-infiltrating cancer cell, a circulating cancer cell, serum, or plasma, and more preferably a cancer resection tissue specimen or a biopsy specimen. In addition, in a case where the sample containing the cancer cell is the cancer resection tissue specimen or the biopsy specimen, these specimens may be subjected to freezing, alcohol fixation, formalin fixation, paraffin wrapping, or a combination of these treatments.
In the present description, "mutation of the amino acid residue" means that a specific amino acid residue in an amino acid sequence of a protein is substituted with an amino acid residue different from an amino acid residue in a corresponding wild-type amino acid sequence. For example, the 326th amino acid residue of the amino acid sequence of the wild-type B-Raf shown in SEQ ID NO: 1 is isoleucine, and substitution of this amino acid residue with an amino acid residue other than isoleucine is called the mutation.
The mutation of the 326th amino acid residue of the amino acid sequence of the B-Raf may be a mutation in which the isoleucine is substituted with phenylalanine, threonine, aspartic acid, lysine, serine, arginine, methionine, glycine, alanine, valine, or leucine. In a case where the mutation of the amino acid residue is the mutation described above, it is possible to select with higher certainty a cancer patient for whom a treatment with the EGFR inhibitor is ineffective. Among these, in a case where the mutation is a mutation (I326V) in which the isoleucine has been substituted with valine, it is possible to select with higher certainty a cancer patient for whom a treatment with the EGFR inhibitor is ineffective.
The determination of the presence or absence of the mutation of the amino acid residue described above may be performed by known methods. The determination of the presence or absence of the mutation may include, for example, detection of a mutation of a base sequence that encodes the 326th amino acid residue of the amino acid sequence of the B-Raf.
In the present description, "mutation of the base sequence" means that at least a part of the bases in the base sequence is substituted with other base such that the amino acid residue encoded by the base sequence becomes different from an amino acid residue encoded by a corresponding wild-type base sequence (also called as a "missense mutation").
The mutation of the base sequence that encodes the 326th amino acid residue of the amino acid sequence of the B-Raf may be a mutation in which the amino acid residue encoded by the base sequence is altered from isoleucine to phenylalanine, threonine, aspartic acid, lysine, serine, arginine, methionine, glycine, alanine, valine, or leucine. In a case where the mutation of the base sequence is the mutation described above, it is possible to select with higher certainty a cancer patient for whom the treatment with the EGFR inhibitor is ineffective. Among these, in a case where the mutation of the base sequence is a mutation in which the base sequence encoding the isoleucine is mutated to a base sequence encoding valine (c.976A>G), it is possible to select with higher certainty a cancer patient for whom the treatment with the EGFR inhibitor is ineffective.
Detection of the mutation of the base sequence may be performed by known methods. The detection of the mutation of the base sequence may be performed by, for example, DNA sequencing, polymerase chain reaction, allele-specific amplification, hybridization using allele-specific probes, mismatch cleavage analysis, single-strand conformation polymorphism, denaturing gradient gel electrophoresis, or temperature gradient gel electrophoresis. The technique may be used alone, or a plurality of techniques may be used in combination.
When the mutation exists on the 326th amino acid residue of the amino acid sequence of the B-Raf of the cancer cell, it is possible to determine that the cancer cell is tolerant to the EGFR inhibitor.
The mechanism between the presence or absence of the mutation of the 326th amino acid residue of the amino acid sequence of the B-Raf of the cancer cell and the tolerance of the cancer cell to the EGFR inhibitor is not certain; however, the inventors of the present invention speculate that the presence or absence of the mutation, and the tolerance of the cancer cell are related to each other at least in the following mechanism. The B-Raf is an activation factor present downstream of the EGFR in an intracellular growth signalling pathway involved in cancer cell growth. The EGFR inhibitor has an effect in which growth signals are inhibited from being transmitted to downstream of the EGFR by inhibiting the function of EGFR, and thus the progression of cancer is suppressed. However, if the mutation exists on the 326th amino acid residue of the amino acid sequence of the B-Raf of the cancer cell, the B-Raf is activated even when the growth signals are not transmitted to downstream of the EGFR due to the EGFR inhibitor, and thus the growth signals are transmitted to downstream of the B-Raf. Therefore, it is considered that, even if the EGFR inhibitor is administered to such a cancer cell, the effect of suppressing the progression of cancer is not exerted, or is unlikely to be exerted.
The present invention may also said to be a method for determining tolerance of the cancer cell to the EGFR inhibitor in a human patient suffering from a cancer, the method including a step of determining the presence or absence of the mutation of the 326th amino acid residue of the amino acid sequence of the B-Raf of the cancer cell by using the sample containing the cancer cell collected from the human patient, wherein the presence of the mutation of the amino acid residue indicate that the cancer cell is tolerant to the anti-EGR antibody drug.
In one embodiment, the cancer cell collected from the cancer patient may have the wild-type KRAS gene. The treatment with the EGFR inhibitor is not effective in some cases even if the cancer cells of the cancer patient have the wild-type KRAS gene. According to this embodiment, by evaluating the efficacy of the EGFR inhibitor in the cancer patient whose cancer cells have the wild-type KRAS gene, it is possible to avoid unnecessary side effects caused by the EGFR inhibitor. In the present description, a wild-type KRAS gene means a KRAS gene having no mutation that gives cancer cells tolerance to the EGFR inhibitor. The mutation is a gene mutation that causes an alteration in the type of 12th and 13th amino acids of the KRAS protein.
In one embodiment, the cancer patient may be a patient who intends to undergo or has undergone a treatment with medication with the EGFR inhibitor. According to this embodiment, it is possible to avoid performing the treatment with medication with the EGFR inhibitor in a patient having cancer cells tolerant to the EGFR inhibitor, or to reduce a dose of the EGFR inhibitor. Accordingly, the side effects caused by the EGFR inhibitor can be avoided or reduced.
In one embodiment, the cancer cell collected from the cancer patient may have a wild-type NRAS gene as well as a wild-type KRAS gene. Generally, a treatment with the EGFR inhibitor is considered to be effective for the cancer patients whose cancer cells have a wild-type NRAS gene as well as wild-type KRAS gene. However, even among such patients, there are patients for whom the treatment with the EGFR inhibitor is ineffective. According to this embodiment, it is possible to discriminate the patients for whom the treatment with the EGFR inhibitor is ineffective among such cancer patients. In the present description, a wild-type NRAS gene means a NRAS gene having no mutation that gives cancer cells tolerance to the EGFR inhibitor. The mutation, for example, is a gene mutation that causes an alteration in the type of a 12th, 13th, 59th, 61st, 117th, or 146th amino acid of the NRAS protein.
In one aspect, the present invention is a method for determining prognosis of cancer, that is a method which involves determining the presence or absence of the mutation of the 326th amino acid residue of the amino acid sequence of the B-Raf of the cancer cell by using the sample containing the cancer cell collected from the human patient suffering from the cancer, wherein the prognosis is determined as poor, when the mutation of the amino acid residue is present.
Hereinbefore, the specific embodiments of the present invention were described in detail, but the present invention is not limited to the above-described embodiments.
Examples
Hereinafter, the present invention will be more specifically described based on examples, but the present invention is not limited to the following examples.
(Detection of Mutation in BRAF Gene)
Patients with a colorectal cancer not having a mutation in codons of the 12th and 13th amino acids of the KRAS protein were treated with a therapy using Panitumumab or Cetuximab (shown in Table 1). A formalin-fixed and paraffin-embedded specimen was prepared from a cancer resection specimen of the patient, and cut into thin sections to prepare two sections with a thickness of 20 μm. A sample obtained by attaching the prepared two sections to a slide glass was used as a sample for DNA extraction. Separately, a sample obtained by preparing a section with a thickness of 4 μm, and attaching the prepared section to a slide glass was used as a sample for microscopic observation. Samples of 26 patients (patients A to Z) were prepared, and mutations in the base sequence of the BRAF gene were detected for each patient sample as follows.
[Table 1]
Patient |
Therapy |
B‑Raf mutation |
Cancer regression index (%) |
A |
IRIS + Panitumumab |
1326V |
115 |
B |
IRIS + Panitumumab |
- |
34.3 |
C |
IRIS + Panitumumab |
- |
18.5 |
D |
IRIS + Panitumumab |
- |
17.8 |
E |
IRIS + Panitumumab |
- |
11.3 |
F |
IRIS + Panitumumab |
D22N |
5 |
G |
FOLFIRI + Cetuximab |
- |
5.1 |
H |
IRIS + Panitumumab CPT - 11 + Cetuximab |
- |
0.1 |
I |
IRIS + Panitumumab |
- |
-7.4 |
J |
CPT - 11 + Cetuximab |
- |
-11.6 |
K |
mFOLFOX6 + Panitumumab |
V600E |
-11.5 |
L |
IRIS + Panitumumab |
- |
-12.5 |
M |
mFOLFOX6 + Cetuximab |
- |
-15.9 |
N |
IRIS + Panitumumab |
- |
-19.1 |
O |
IRIS + Panitumumab mFOLFOX6 + Cetuximab sLV5FU2 + Cetuximab |
N581Y |
-22.4 |
P |
IRIS + Panitumumab |
- |
-27 |
Q |
FOLFIRI + Cetuximab |
- |
-26.6 |
R |
CPT - 11 + Cetuximab |
- |
-32.8 |
S |
mFOLFOX6 + Panitumumab |
- |
-33.8 |
T |
IRIS + Panitumumab |
- |
-33.7 |
U |
CPT - 11 + Panitumumab |
V600E |
-35.7 |
V |
CPT - 11 + Panitumumab |
- |
-36.8 |
W |
IRIS + Panitumumab |
- |
-52.8 |
X |
mFOLFOX6 + Cetuximab |
- |
-55 |
Y |
mFOLFOX6 + Panitumumab |
- |
-72.3 |
Z |
IRIS + Panitumumab |
- |
-72.7 |
The sample for microscopic observation was stained with haematoxylin and eosin. The sample after the staining was observed with a microscope and a site containing many cancer cells in the section was specified. From the sample for DNA extraction, the site specified in the sample for microscopic observation was scraped with a razor, and DNA was extracted from the scraped site. The DNA extraction was carried out by using BiOstic FFPE Tissue DNA Isolation Kit (trade name).
The BRAF gene was isolated from the DNA of the specimen by using, as a probe, a probe nucleic acid containing a part or all of continuous sequences (100 to 130 bases) in each of 18 exon sequences of the BRAF gene, or containing a complementary sequence thereof. The base sequence of the BRAF gene was analysed by MiSeq sequencer of Illumina, Inc. to detect mutations in the base sequence of the BRAF gene.
For the patient A, the mutation (c.976A>G) that causes the mutation of I326V in the amino acid sequence of the B-Raf was detected in the base sequence of the BRAF gene. For the patients F, K, O, und U, mutations that cause mutations of D22N, V600E, N581Y, and V600E, respectively, were detected in the amino acid sequence of the B-Raf in the base sequence of the BRAF gene. For the other patients, no mutation that causes a mutation in the amino acid sequence of the B-Raf was detected in the base sequence of the BRAF gene.
(Effect of Treatment with Anti-EGFR Antibody Drug)
For the patients A to Z, a cancer regression index (%) was calculated as the following formula:
Cancer regression index (%) = (diameter of primary lesion after treatment + diameter of metastatic lesion after treatment) / (diameter of primary lesion before treatment + diameter of metastatic lesion before treatment) x 100 - 100
The relationship between the presence or absence of the mutation of the BRAF gene, and the effect of the treatment with the anti-EGFR antibody drug in the patients A to Z is shown in Fig. 1 and Tables 1 and 2.
[Table 2]
B‑Raf mutation |
Number of patients |
Average cancer regression index (%) |
Standard deviation |
1326V |
1 |
115 |
- |
D22N V600E N581Y |
4 |
-16.3 |
17.3 |
No mutation |
22 |
-19 |
28.9 |
As shown in Fig. 1 and Tables 1 and 2, in the patient A for whom the mutation that causes the mutation in the 326th amino acid residue of the amino acid sequence of the B-Raf was detected in the base sequence of the BRAF gene, the progression of cancer was 10 recognised, and the cancer cells of the patient A had tolerance to the anti-EGFR antibody drug.
SEQUENCE LISTING




Claims
1. A method for determining tolerance of a cancer cell to an epidermal growth factor receptor inhibitor in a human patient suffering from a cancer, the method comprising:
R. 43(1)(a)
Independent claim
a step of determining the presence or absence of a mutation of a 326th amino acid residue of an amino acid sequence of a B-Raf protein of the cancer cell by using a sample containing the cancer cell collected from the human patient,
wherein the cancer cell is determined as tolerant to the epidermal growth factor receptor inhibitor, when the mutation of the amino acid residue is present.
wherein the epidermal growth factor receptor inhibitor is an anti-epidermal growth factor receptor antibody drug.
3. The method according to claim 1 or 2,
wherein the determination of the presence or absence of the mutation of the amino acid residue includes detection of a mutation of a base sequence encoding the 326th amino acid residue of the amino acid sequence of the B-Raf protein.
4. The method according to claim 3,
wherein the detection of the mutation of the base sequence is performed by DNA sequencing, polymerase chain reaction, allele-specific amplification, hybridization using allele-specific probes, mismatch cleavage analysis, single-strand conformation polymorphism, denaturing gradient gel electrophoresis, or temperature gradient gel electrophoresis.
5. The method according to-any one of claims 1 to 4,
wherein the sample is a cancer resection tissue specimen, a biopsy specimen, an ascites-infiltrating cancer cell, a circulating cancer cell, serum, or plasma.
6. The method according to any one of claims 1 to 5,
wherein the cancer is a colorectal cancer or a rectal cancer.
7. The method according to any one of claims 1 to 6,
wherein the mutation of the amino acid residue is I326V.
8. The method according to claim 3 or 4,
wherein the mutation of the base sequence is c.976A>G.
9. The method according to any one of claims 1 to 8,
wherein the cancer cell has a wild-type KRAS gene.
Abstract
METHOD FOR DETERMINING TOLERANCE OF CANCER CELL TO EPIDERMAL GROWTH FACTOR RECEPTOR INHIBITOR
R. 47(1)
Title of invention
The present invention provides a method for determining tolerance of a cancer cell to an EGFR inhibitor in a human patient suffering from a cancer, the method comprising a step of determining the presence or absence of a mutation of a 326th amino acid residue of an amino acid sequence of a B-Raf protein of the cancer cell by using a sample containing the cancer cell collected from the human patient, wherein the cancer cell is determined as tolerant to the epidermal growth factor receptor inhibitor, when the mutation of the amino acid residue is present. According to such a method, it is possible to select in advance a cancer patient for whom a treatment with the EGFR inhibitor is ineffective.